Polyploidy
polyploidy as a pathway to speciation and key innovations
Local adaptation, speciation, and the drivers of ecosystem change
Polyploidy was long appreciated in plants, but took on a new importance since the advent of genomics as it simultaneously became a source of novelty and a challenge to many assumptions of molecular evolution derived from diploids. The lab is currently focused on neopolyploids and how increases in ploidy within a species can lead to niche diversfication and local adaptation to future climate change conditions. While organismally focused in a few grass lineages, the lab is interesting in developing methods that facilitate population genomic analyses of polyploids 1 and several packages and pipelines are underway. By using some widely-distributed species as models, the lab aims to pair the large-scale inferential power of population genomics with field and controlled experiments to demonstrate expected versus observed effects of polyploidy on traits such as drought tolerance.
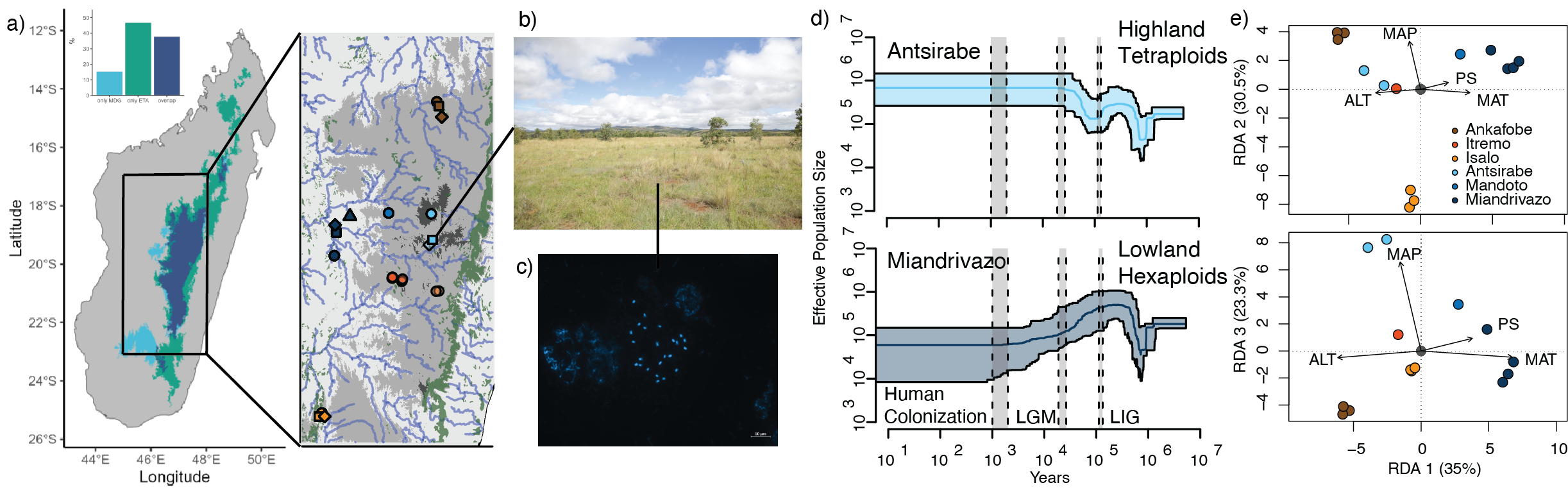 Unpublished figure from Tiley et al. in prep Expected distribution of Loudetia simplex in Madagasscar and demographic histories of tetraploids versus hexaploids. Linear discriminate analyses show associations between environment and ploidy.
Unpublished figure from Tiley et al. in prep Expected distribution of Loudetia simplex in Madagasscar and demographic histories of tetraploids versus hexaploids. Linear discriminate analyses show associations between environment and ploidy.
Ancient whole-genome duplication and diversification
Previously, ancient whole-genome duplications (WGDs) have also been a priority with several methods for detecting ancient WGDs developed that use distributions of evolutionary distances between paralogs within a species 2, distributions of gene count data accompanied by a phylogeny 3, or gene trees 4 to place WGDs in a phylogenetic context. The lab remains broadly interested in the consequences of polyploidy on genome architecture and gene family evolution and new projects that investigate the evolutionary contributions of ancient WGDs are expected. Polyploidy in the context of allpolyploid complexes are also a topic of interest and this led to a pipeline for phasing homeologous short-read data 5. Phased data can help include poplyploids in population genomics investigations, but there are still a number of assumptions regarding auto- and allopolyploids to resolve.
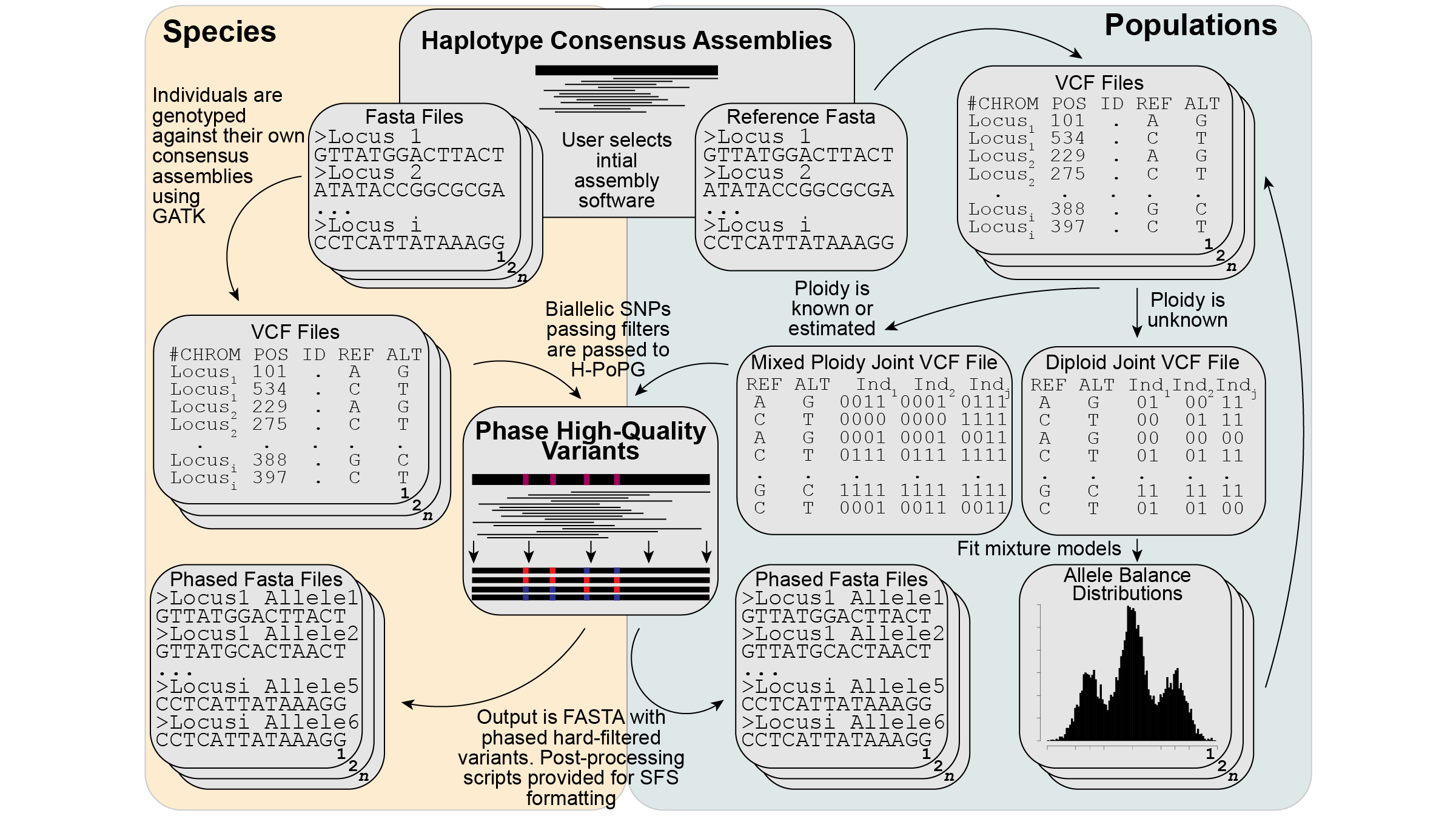 Figure 2 from Tiley et al. 2024 5 Pipeline of mapping reads back to consensus loci and binning phasing subproblems to make the computation tractable. It is possible to treat all individuals as there own reference when working with a collection of species or to use a sinlge reference when working with populations pf the same or similar species.
Figure 2 from Tiley et al. 2024 5 Pipeline of mapping reads back to consensus loci and binning phasing subproblems to make the computation tractable. It is possible to treat all individuals as there own reference when working with a collection of species or to use a sinlge reference when working with populations pf the same or similar species.
-
Tiley GP, Crowl AA, Almary TOM, Luke WRQ, Solofondranohatra CL, Besnard G, Lehmann CER, Yoder AD, Vorontsova MS. 2023. Genetic variation in Loudetia simplex supports the presence of ancient grasslands in Madagascar. Plants People Planet 6:315-329 ↩
-
Tiley GP, Barker MS, Burleigh JG. 2018. Assessing the Performance of Ks Plots for Detecting Ancient Whole Genome Duplications. Genome Biology and Evolution 10:2882-2898. ↩
-
Tiley GP, Ané C, Burleigh JG. 2016. Evaluating and characterizing ancient whole-genome duplications in plants with gene count data. Genome Biology and Evolution 8:1023-1037. ↩
-
Li Z†,Tiley GP†, Galuska SR, Reardon CR, Kidder TI, Rundell RJ, Barker MS. 2018. Multiple large-scale gene and genome duplications during the evolution of hexapods. PNAS 115:4713-4718. ↩
-
Tiley GP, Crowl AA, Manos PS, Sessa EB, Solís-Lemus C, Yoder AD, Burleigh JG. 2024. Benefits and Limits of Phasing Alleles for Network Inference of Allopolyploid Complexes. Systematic Biology syae024. ↩ ↩2