Plant Phylogenomics
using phylogenetics and molecular evolution to understand 450 million years of diversity
Phylogenetic Networks
The lab is primarily interested in phylogenomic networks. While reticulate evolution has long been recognized as a feature of plants, our ability to represent molecular evolution as a network rather than a tree is relatively new and transformative 1. Previous work includes estimating the age of hybridization events 2, applying networks to polyploid complexes 3, and prooving the statistical identifiability of networks under various criteria 4. The lab continues to work on these areas as well as the empirical application of networks to explaining genealogical discordance throughout the plant Tree of Life and more specifically grasses. Some models for estimating networks from polyploid complexes and testing hypotheses of trait evolution are also under development.
Estimating and dating the plant Tree of Life
There are also a number of empirical and theoretical investigations into divergence time estimation and variation in rates of evolution across lineages and loci. Although there is a long legacy of divergence time studies in plants, it is only recent that representative chromosome-level genomes have become available to understand the importance of genome structure in rate variation. Some divergence time estimates are also worth revisiting with large multi-locus nuclear data as previous work has shown how biases due to incomplete lineage sorting can propogate throughout the tree of life 5.
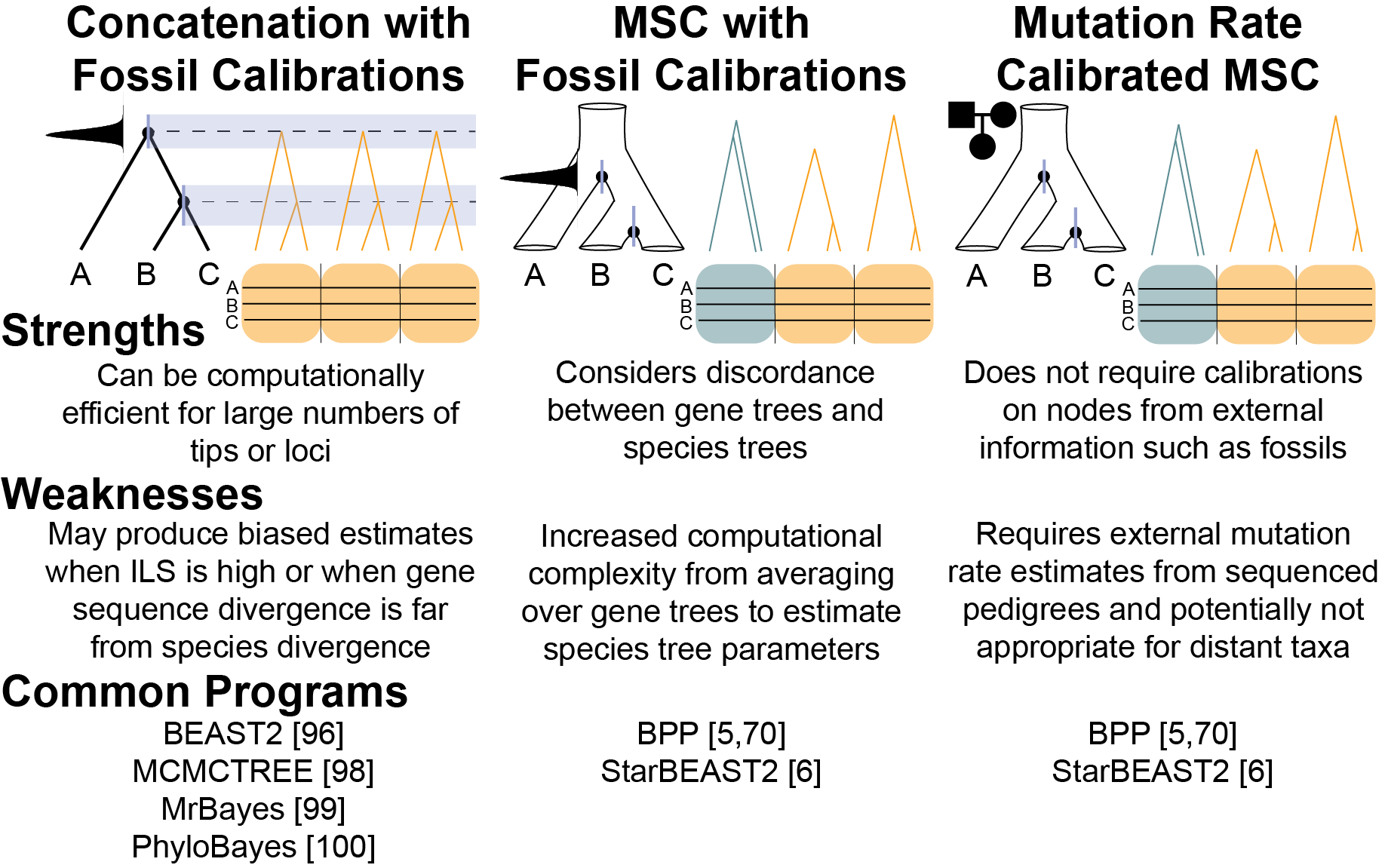 Figure 5 from Tiley et al. 2020 5 Comparison of different divergence time estimation strategies.
Figure 5 from Tiley et al. 2020 5 Comparison of different divergence time estimation strategies.
Previous research in this area included participating in large-scale phylogenomic efforts 6,7 and biodiversity genomics 8 that not only seek to investigate phylogenetic relationships, but also build the necessary resources for the scientific community to rigorously test phylogenetic hypotheses. Such data have been used to understand general features of plant genome evolution such as the evolution of recombination rate and transposable element content 9. Future directions could also combine comparative genomics with phylogenetic models to reveal broad patterns and processes of plant genome evolution associated with interesting traits.
-
Kong S, Solís-Lemus C, Tiley GP. 2025. Phylogenetic networks empower biodiversity science. PNAS 122:e2410934122. doi: https://doi.org/10.1073/pnas.2410934122 ↩
-
Tiley GP, Flouri T, Jiao X, Poelstra JW, Xu B, Zhu T, Rannala B, Yoder AD, Yang Z. 2023. Estimation of species divergence times in presence of cross-species gene flow. Systematic Biology 72:820-836. ↩
-
Tiley GP, Crowl AA, Manos PS, Sessa EB, Solís-Lemus C, Yoder AD, Burleigh JG. 2024. Benefits and Limits of Phasing Alleles for Network Inference of Allopolyploid Complexes. Systematic Biology syae024. ↩
-
Tiley GP, Solís-Lemus C. 2023. Extracting diamonds: Identifiability of 4-node cycles in level-1 phylogenetic networks under a pseudolikelihood coalescent model. bioRxiv doi: https://doi.org/10.1101/2023.10.25.564087 ↩
-
Tiley GP, Poelstra JW, dos Reis M, Yang Z, Yoder AD. 2020. Molecular Clocks without Rocks: New Solutions for Old Problems. Trends in Genetics 36:845-856. ↩ ↩2
-
Leebens-Mack et al. 2019. [One thousand plant transcriptomes and the phylogenomics of green plants.x]x(https://www.nature.com/articles/s41586-019-1693-2) Nature 574:679-685. ↩
-
Breinholt JW, Carey SB, Tiley GP, Davis EC, Endara L, McDaniel SF, Neves LG, Sessa EB, von Konrat M, Chantanaorrapint S, Fawcett S, Ickert-Bond SM, Labiak PH, Larraín J, Lehnert M, Lewis LR, Nagalingum NS, Patel N, Rensing SA, Testo W, Vasco A, Villarreal JC, Williams EW, Burleigh JG. 2021. A target enrichment probe set for resolving the flagellate plant tree of life. Applications in Plant Sciences 9:e11406. ↩
-
Carey SB, Jenkins J, Payton AC, Shu S, Lovell JT, Maumus F, Sreedasyam A, Tiley GP, Fernandez-Pozo N, Barry K, Chen C, Wang M, Lipzen A, Daum C, Saski CA, McBreen JC, Conrad RE, Kollar LM, Olsson S, Huttunen S, Landis JB, Burleigh JB, Wickett NJ, Johnson MG, Rensing SA, Grimwood J, Schmutz J, McDaniel SF. 2021. Chromosome fusions shape an ancient UV sex chromosome system. Science Advances 7:eabh2488. ↩
-
Tiley GP, Burleigh JG. 2015. The relationship of recombination rate, genome structure, and patterns of molecular evolution across angiosperms. BMC Evolutionary Biology 15:194. ↩